Difference between revisions of "Research/key-initiatives/ras/target-identification/structural-biology"
(Marked this version for translation) |
|||
| Line 1: | Line 1: | ||
<languages/> | <languages/> | ||
<translate> | <translate> | ||
| − | == RAS Structural Biology == | + | == RAS Structural Biology == <!--T:1--> |
| + | <!--T:2--> | ||
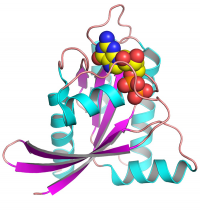
[[File:Ras-structural-biology-article.png|200px|thumb|right|Structure of wild-type KRAS4b in the GDP-bound form]] | [[File:Ras-structural-biology-article.png|200px|thumb|right|Structure of wild-type KRAS4b in the GDP-bound form]] | ||
| + | <!--T:3--> | ||
Recent studies from various laboratories have renewed our hope for the development of RAS-inhibitory molecules. Our group leads the structural biology efforts within the RAS Initiative. Our aim is to gain structural insights into wild-type and oncogenic mutants of KRAS in complex with various effectors/regulatory/partner proteins, which may identify novel binding pockets or interfaces amenable to attack with small molecules. | Recent studies from various laboratories have renewed our hope for the development of RAS-inhibitory molecules. Our group leads the structural biology efforts within the RAS Initiative. Our aim is to gain structural insights into wild-type and oncogenic mutants of KRAS in complex with various effectors/regulatory/partner proteins, which may identify novel binding pockets or interfaces amenable to attack with small molecules. | ||
| − | === Our Progress === | + | === Our Progress === <!--T:4--> |
| + | <!--T:5--> | ||
Our group has solved the first structure of KRAS4b protein in complex with PDEdelta, a protein that plays an important role in targeting KRAS4b to cellular membranes. This protein-protein complex structure shows every amino acid of full-length, fully processed KRAS protein for the first time. | Our group has solved the first structure of KRAS4b protein in complex with PDEdelta, a protein that plays an important role in targeting KRAS4b to cellular membranes. This protein-protein complex structure shows every amino acid of full-length, fully processed KRAS protein for the first time. | ||
| + | <!--T:6--> | ||
Working to gain structural insights into KRAS mutations, we have solved the first structures of multiple oncogenic mutants of KRAS4b in the GTP-bound form. The GTP-bound forms of these mutants are the most common drivers of human cancers. | Working to gain structural insights into KRAS mutations, we have solved the first structures of multiple oncogenic mutants of KRAS4b in the GTP-bound form. The GTP-bound forms of these mutants are the most common drivers of human cancers. | ||
| − | === Our Projects === | + | === Our Projects === <!--T:7--> |
| + | <!--T:8--> | ||
*Solve structures of wild-type and oncogenic mutants of KRAS in the active state | *Solve structures of wild-type and oncogenic mutants of KRAS in the active state | ||
| + | <!--T:9--> | ||
*Determine structures of KRAS complexes with various effectors and regulatory/trafficking proteins to aid structure- | *Determine structures of KRAS complexes with various effectors and regulatory/trafficking proteins to aid structure- | ||
based drug design | based drug design | ||
| + | <!--T:10--> | ||
*Exploit structures for virtual compound screening followed by biochemical, biophysical and structural studies | *Exploit structures for virtual compound screening followed by biochemical, biophysical and structural studies | ||
| − | === Tools We Use === | + | === Tools We Use === <!--T:11--> |
| + | <!--T:12--> | ||
*Nanoliter-scale protein crystallization system | *Nanoliter-scale protein crystallization system | ||
| + | <!--T:13--> | ||
*Automated protein crystallization imaging system | *Automated protein crystallization imaging system | ||
| + | <!--T:14--> | ||
*Synchrotron beam line | *Synchrotron beam line | ||
| + | <!--T:15--> | ||
*In-house X-ray diffractometer | *In-house X-ray diffractometer | ||
| + | <!--T:16--> | ||
*Isothermal titration calorimetry | *Isothermal titration calorimetry | ||
| + | <!--T:17--> | ||
*Farnesylated and methylated KRAS4b | *Farnesylated and methylated KRAS4b | ||
| − | === Collaborations === | + | === Collaborations === <!--T:18--> |
| + | <!--T:19--> | ||
The RAS Structural Biology Group has collaborated with: | The RAS Structural Biology Group has collaborated with: | ||
| + | <!--T:20--> | ||
Nir London | Nir London | ||
| + | <!--T:21--> | ||
Weizmann Institute of Science, Rehovot, Israel | Weizmann Institute of Science, Rehovot, Israel | ||
| + | <!--T:22--> | ||
Hans Robert Kalbitzer | Hans Robert Kalbitzer | ||
University of Regensburg, Germany | University of Regensburg, Germany | ||
| + | <!--T:23--> | ||
Arul Chinnaiyan | Arul Chinnaiyan | ||
Michigan Center for Translational Pathology, Ann Arbor, MI | Michigan Center for Translational Pathology, Ann Arbor, MI | ||
| − | === Contact === | + | === Contact === <!--T:24--> |
| + | <!--T:25--> | ||
[[File:Dhirendra-simanshu-article.jpg|200px|thumb|right|Dr. Dhirendra Simanshu, Structural Biology Group Lead]] | [[File:Dhirendra-simanshu-article.jpg|200px|thumb|right|Dr. Dhirendra Simanshu, Structural Biology Group Lead]] | ||
| + | <!--T:26--> | ||
For more information, contact the RAS Structural Biology Group team lead: | For more information, contact the RAS Structural Biology Group team lead: | ||
| + | <!--T:27--> | ||
'''Dr. Dhirendra Simanshu''' | '''Dr. Dhirendra Simanshu''' | ||
| + | <!--T:28--> | ||
301-360-3438 | 301-360-3438 | ||
| + | <!--T:29--> | ||
dhirendra.simanshu@nih.gov | dhirendra.simanshu@nih.gov | ||
</translate> | </translate> | ||
Latest revision as of 21:48, 29 October 2019
Contents
RAS Structural Biology
Recent studies from various laboratories have renewed our hope for the development of RAS-inhibitory molecules. Our group leads the structural biology efforts within the RAS Initiative. Our aim is to gain structural insights into wild-type and oncogenic mutants of KRAS in complex with various effectors/regulatory/partner proteins, which may identify novel binding pockets or interfaces amenable to attack with small molecules.
Our Progress
Our group has solved the first structure of KRAS4b protein in complex with PDEdelta, a protein that plays an important role in targeting KRAS4b to cellular membranes. This protein-protein complex structure shows every amino acid of full-length, fully processed KRAS protein for the first time.
Working to gain structural insights into KRAS mutations, we have solved the first structures of multiple oncogenic mutants of KRAS4b in the GTP-bound form. The GTP-bound forms of these mutants are the most common drivers of human cancers.
Our Projects
- Solve structures of wild-type and oncogenic mutants of KRAS in the active state
- Determine structures of KRAS complexes with various effectors and regulatory/trafficking proteins to aid structure-
based drug design
- Exploit structures for virtual compound screening followed by biochemical, biophysical and structural studies
Tools We Use
- Nanoliter-scale protein crystallization system
- Automated protein crystallization imaging system
- Synchrotron beam line
- In-house X-ray diffractometer
- Isothermal titration calorimetry
- Farnesylated and methylated KRAS4b
Collaborations
The RAS Structural Biology Group has collaborated with:
Nir London
Weizmann Institute of Science, Rehovot, Israel
Hans Robert Kalbitzer University of Regensburg, Germany
Arul Chinnaiyan Michigan Center for Translational Pathology, Ann Arbor, MI
Contact
For more information, contact the RAS Structural Biology Group team lead:
Dr. Dhirendra Simanshu
301-360-3438
dhirendra.simanshu@nih.gov