Difference between revisions of "Research/key-initiatives/ras/screens-assays/drug-discovery"
(Marked this version for translation) |
|||
| Line 1: | Line 1: | ||
<languages/> | <languages/> | ||
<translate> | <translate> | ||
| − | == RAS Drug Discovery == | + | == RAS Drug Discovery == <!--T:1--> |
| + | <!--T:2--> | ||
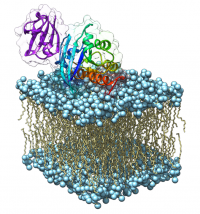
[[File:Ras-drug-discovery-article.png|200px|thumb|right|Model of a RAS-RAF complex on a membrane]] | [[File:Ras-drug-discovery-article.png|200px|thumb|right|Model of a RAS-RAF complex on a membrane]] | ||
| + | <!--T:3--> | ||
The goal of the Drug Discovery group is to develop assays that will measure aspects of RAS biology upon which human cancer cells depend. These assays comprise both phenotypic (i.e., cell-based) and molecular (in vitro) screens. | The goal of the Drug Discovery group is to develop assays that will measure aspects of RAS biology upon which human cancer cells depend. These assays comprise both phenotypic (i.e., cell-based) and molecular (in vitro) screens. | ||
| + | <!--T:4--> | ||
The cell-based screens developed utilizing RAS-dependent cell lines created by Mariano Barbacid, CNIO, Madrid, allow selection of molecules that preferentially inhibit cells expressing mutant RAS. Our in vitro screens focus on: | The cell-based screens developed utilizing RAS-dependent cell lines created by Mariano Barbacid, CNIO, Madrid, allow selection of molecules that preferentially inhibit cells expressing mutant RAS. Our in vitro screens focus on: | ||
| + | <!--T:5--> | ||
*KRAS-lipid interactions, | *KRAS-lipid interactions, | ||
| + | <!--T:6--> | ||
*KRAS-effector protein-protein interactions, and | *KRAS-effector protein-protein interactions, and | ||
| + | <!--T:7--> | ||
*KRAS dependent activation of effector pathways. | *KRAS dependent activation of effector pathways. | ||
| + | <!--T:8--> | ||
Successful assay formats are being made available to academic and commercial organizations for use in high-throughput screening programs that we hope will yield drugs useful in treating human cancers. | Successful assay formats are being made available to academic and commercial organizations for use in high-throughput screening programs that we hope will yield drugs useful in treating human cancers. | ||
| − | === Our Progress === | + | === Our Progress === <!--T:9--> |
| + | <!--T:10--> | ||
Our group has developed and validated assays suitable for high-throughput screening of libraries of compounds to discover tool molecules and drug candidates. | Our group has developed and validated assays suitable for high-throughput screening of libraries of compounds to discover tool molecules and drug candidates. | ||
| + | <!--T:11--> | ||
Our in vitro biochemical assays are qualified for finding molecules that affect RAS-effector and RAS-lipid interactions, as well as those that affect GTP hydrolysis. | Our in vitro biochemical assays are qualified for finding molecules that affect RAS-effector and RAS-lipid interactions, as well as those that affect GTP hydrolysis. | ||
| + | <!--T:12--> | ||
Our group’s cell-based assays are validated for screening based on RAS-dependent growth, RAS effector (MEK/ERK) activation, RAS-effector binding, and RAS localization. | Our group’s cell-based assays are validated for screening based on RAS-dependent growth, RAS effector (MEK/ERK) activation, RAS-effector binding, and RAS localization. | ||
| − | === Our Projects === | + | === Our Projects === <!--T:13--> |
| + | <!--T:14--> | ||
*High-throughput screens of small molecule libraries | *High-throughput screens of small molecule libraries | ||
| + | <!--T:15--> | ||
*Validation of hit/lead molecules with secondary screens | *Validation of hit/lead molecules with secondary screens | ||
| + | <!--T:16--> | ||
*Synthesis and testing of lead compounds to improve potency | *Synthesis and testing of lead compounds to improve potency | ||
| + | <!--T:17--> | ||
*Transfer of screening technologies to academic and industrial partners | *Transfer of screening technologies to academic and industrial partners | ||
| − | === Tools We Use === | + | === Tools We Use === <!--T:18--> |
| + | <!--T:19--> | ||
*Mouse embryonic fibroblasts engineered to be dependent on single alleles of RAS | *Mouse embryonic fibroblasts engineered to be dependent on single alleles of RAS | ||
| + | <!--T:20--> | ||
*Tool compounds and approved drugs | *Tool compounds and approved drugs | ||
| + | <!--T:21--> | ||
*Nanodiscs | *Nanodiscs | ||
| + | <!--T:22--> | ||
*Fully-processed KRAS protein | *Fully-processed KRAS protein | ||
| − | === Collaborations === | + | === Collaborations === <!--T:23--> |
| + | <!--T:24--> | ||
The Drug Discovery Group has collaborated with: | The Drug Discovery Group has collaborated with: | ||
Debbie Morrison | Debbie Morrison | ||
National Cancer Institute | National Cancer Institute | ||
| + | <!--T:25--> | ||
Karla Satchell | Karla Satchell | ||
Northwestern University | Northwestern University | ||
| + | <!--T:26--> | ||
Daiichi Sankyo | Daiichi Sankyo | ||
Sanofi-Aventis | Sanofi-Aventis | ||
| + | <!--T:27--> | ||
William Garland | William Garland | ||
Tosk, Inc. | Tosk, Inc. | ||
| + | <!--T:28--> | ||
Cameron Pitt | Cameron Pitt | ||
KyRAS Therapeutics | KyRAS Therapeutics | ||
| − | === Contact === | + | === Contact === <!--T:29--> |
| + | <!--T:30--> | ||
For more information about RAS Drug Discovery, contact Jim Hartley, james.hartley@nih.gov. | For more information about RAS Drug Discovery, contact Jim Hartley, james.hartley@nih.gov. | ||
</translate> | </translate> | ||
Latest revision as of 21:48, 29 October 2019
Contents
RAS Drug Discovery
The goal of the Drug Discovery group is to develop assays that will measure aspects of RAS biology upon which human cancer cells depend. These assays comprise both phenotypic (i.e., cell-based) and molecular (in vitro) screens.
The cell-based screens developed utilizing RAS-dependent cell lines created by Mariano Barbacid, CNIO, Madrid, allow selection of molecules that preferentially inhibit cells expressing mutant RAS. Our in vitro screens focus on:
- KRAS-lipid interactions,
- KRAS-effector protein-protein interactions, and
- KRAS dependent activation of effector pathways.
Successful assay formats are being made available to academic and commercial organizations for use in high-throughput screening programs that we hope will yield drugs useful in treating human cancers.
Our Progress
Our group has developed and validated assays suitable for high-throughput screening of libraries of compounds to discover tool molecules and drug candidates.
Our in vitro biochemical assays are qualified for finding molecules that affect RAS-effector and RAS-lipid interactions, as well as those that affect GTP hydrolysis.
Our group’s cell-based assays are validated for screening based on RAS-dependent growth, RAS effector (MEK/ERK) activation, RAS-effector binding, and RAS localization.
Our Projects
- High-throughput screens of small molecule libraries
- Validation of hit/lead molecules with secondary screens
- Synthesis and testing of lead compounds to improve potency
- Transfer of screening technologies to academic and industrial partners
Tools We Use
- Mouse embryonic fibroblasts engineered to be dependent on single alleles of RAS
- Tool compounds and approved drugs
- Nanodiscs
- Fully-processed KRAS protein
Collaborations
The Drug Discovery Group has collaborated with: Debbie Morrison National Cancer Institute
Karla Satchell Northwestern University
Daiichi Sankyo Sanofi-Aventis
William Garland Tosk, Inc.
Cameron Pitt KyRAS Therapeutics
Contact
For more information about RAS Drug Discovery, contact Jim Hartley, james.hartley@nih.gov.